In the early days of natural language processing (NLP) research, we quantified the similarity between documents by counting shared words. However, this simplistic approach ignored synonyms, treating “cat” and “kitty” as distinct entities, much like “cat” and “rock.” (Figure 1. Left) The resulting identity-based word representation, known as one-hot encoding, fell short when applied to clustering documents. Fortunately, the NLP landscape has evolved, thanks to deep learning techniques like Word2Vec [1]. “cat” and “kitty” — once distant, now recognized as near identical due to Word2Vec (Figure 1. Right).

We faced a similar dilemma in gene analysis. When comparing gene lists, we counted shared genes, akin to the old NLP approach. However, genes like TLR7 and MYD88, with remarkably similar biological roles in innate immune signaling, were missed when appearing in separate lists. Just as “cat” and “kitty” were once distinct and unrecognized. In our recent Nature Communications publication [2], we draw parallels between NLP and bioinformatics, introducing Functional Representation of Gene Signatures (FRoGS) as the Word2Vec equivalent for gene analysis (Figure 2). We then harnessed the power of FRoGS to uncover compound targets. By aligning gene signatures from shRNA/cDNA perturbations with compound treatment data from the Broad’s L1000 dataset, we achieved remarkable results. Specifically, FRoGS-based AI models demonstrated a 36% recall in identifying true compound targets, surpassing the meager 9% recall achieved using the traditional one-hot encoding approach.
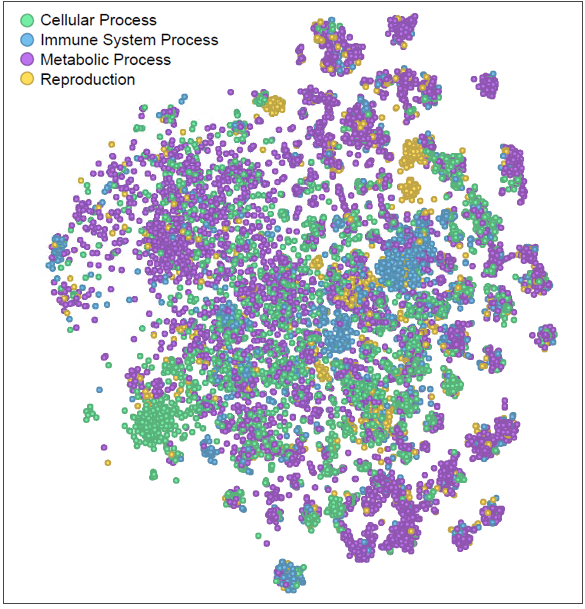
Why does FRoGS work? Gene functions lie at the heart of biological processes and phenotypic outcomes. When genes are naively represented as one-hot vectors, AI models must painstakingly rediscover their functions—a process demanding copious amounts of training data. Unfortunately, such extensive data may not always be available, hindering accurate predictions. FRoGS embeddings revolutionize gene representation. Each human gene is encoded as a meaningful vector, capturing both its known functions annotated in Gene Ontology (GO) [3] and its latent roles inferred from large-scale transcriptome datasets (such as ARCHS4) [4]. FRoGS enables AI models to focus on learning the intricate associations between gene functions and phenotypes without rediscovering gene functions.
In this blog we further demonstrate how FRoGS vectors enable powerful machine learning tasks, revolutionizing gene analysis beyond traditional one-hot encoding.
Example 1: Tissue-Specific Gene Expression
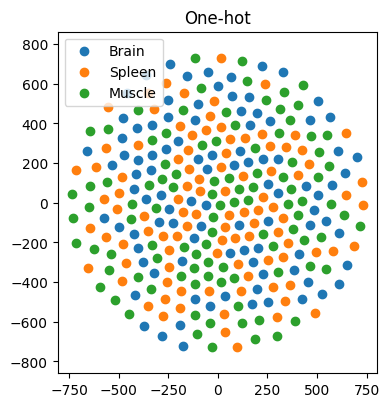
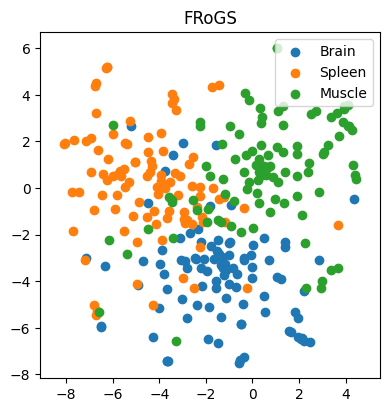
In our first example, we work with 97, 98, and 100 genes specifically expressed in the brain, spleen, and muscle, respectively. Our goal is to predict the tissue of gene expression. Traditionally, one-hot encoding places these genes in a high-dimensional space, with no inherent similarity. Consequently, AI models struggle to learn meaningful patterns, resulting in dismal classification accuracy of approximately 29% (±3%) (n = 100 simulations) — akin to random guessing (Figure 3). However, when we represent genes using FRoGS vectors, tissue-specific clusters emerge in t-SNE plots (Figure 4). The random forest accuracy soars to approximately 80% (±5%) (n = 100 simulations), demonstrating the power of FRoGS even with limited data (Figure 4).
Example 2: Gene Lists and Functional Signatures
Our dataset comprises 35, 24, and 122 gene lists associated with artery, heart, and brain, respectively. Each gene list consists of approximately 100 gene members. In the one-hot encoding approach, a gene list is represented as the sum of its constituent one-hot member gene vectors. While these gene lists exhibit some clustering patterns (Figure 5), they pale in comparison to the pronounced clusters formed by FRoGS gene signature embeddings (Figure 6). As a result, the classification accuracy for prediction is 85% (±4%) (n = 100 simulations) for one-hot vectors and 100.0% (±0.4%) (n = 100 simulations) for FRoGS vectors.


Conclusion
FRoGS heralds a new era in bioinformatics, akin to Word2Vec’s impact on NLP. As researchers, let’s embrace this advancement and explore FRoGS’ potential across diverse biological contexts. Visit our GitHub repository [5] and unlock the transformative capabilities of FRoGS in your next machine learning endeavor.
References