Coronascape is a data hub depositing public available COVID-19 research related omics data sets. It includes more than 200 processed gene lists for SARS-CoV-2 from more than 20 studies. These gene lists were generated using several omics technologies, including transcriptome (RNA-Seq and scRNASeq), proteome, phosphoproteome, ubiquitome, and interactome, providing a comprehensive picture of SARS-CoV-2 infection in various host cell and tissue types. All data sets deposited in Coronascape were retrieved from published scientific articles and data sets. If publicly available, the gene lists were downloaded from supplementary tables in the publications, then processed according to the criteria suggested by the publications. Coronascape has a friendly simple user interface and convenient search function for different SARS-CoV-2 omics data sets. Users can search for similar gene lists from public data sets compared to user provided gene list, and perform system level analysis for the selected multiple gene lists using Metascape. Coupled with Metascape, Coronascape provides quick access to numerous published COVID-19 omics data sets, and a comprehensive system level data analysis toolkit for data mining.
Comma separated gene symbols or any other types of IDs supported by Metascape. You can click try example to test an example list of gene symbols.
Use the below functions to add gene lists from public data sets for Metascape Analysis
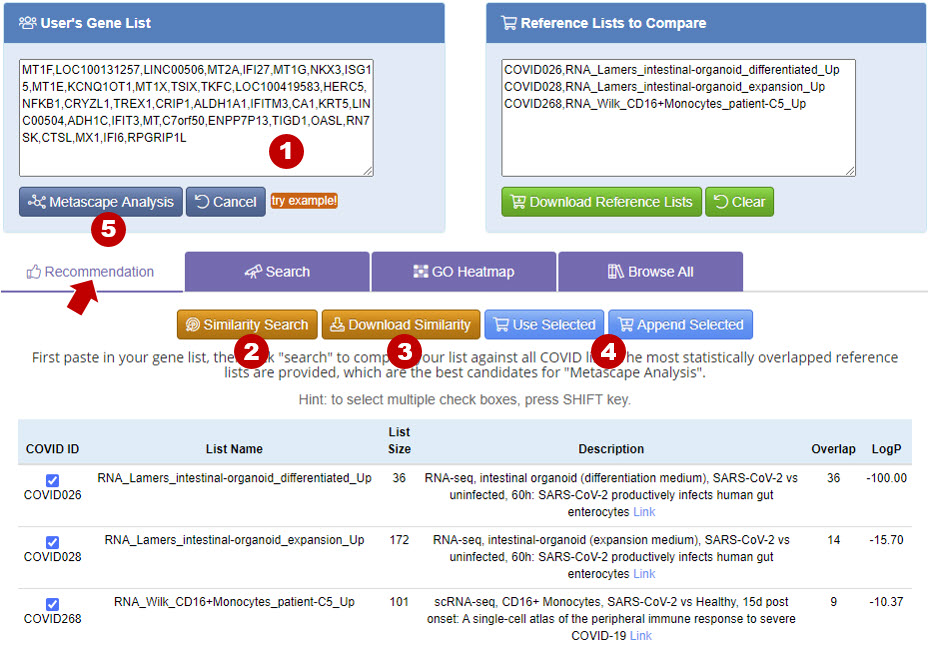
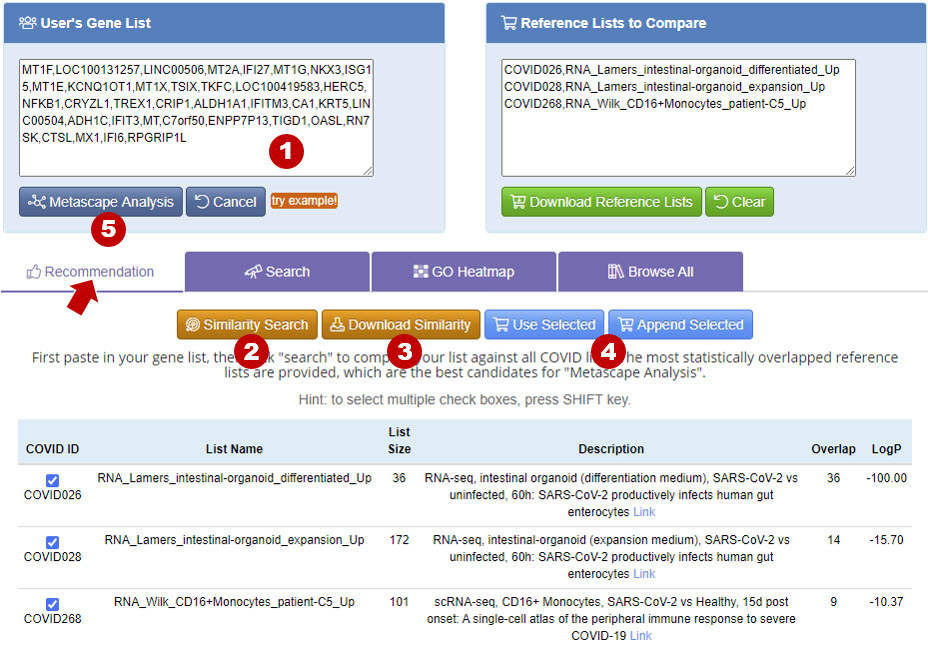
Recommendation provides Similarity Search function to compare the user provided gene list to the gene lists deposited in Coronascape. After user uploads a gene list using User’s Gene List panel, click Similarity Search, then you will see the results. Click Download Similarity will download the similarity score and genes overlapping with your list to text file. By clicking Use Selected or Append Selected button, you can add the selected gene lists to the Reference Lists to Compare panel, and perform Metascape Analysis for the lists.
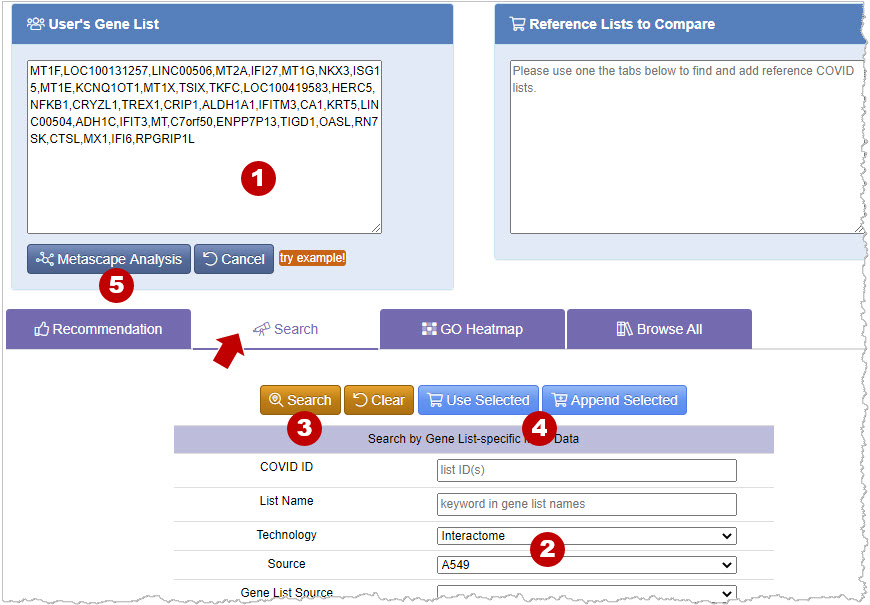
Search function provide a convenient way to search for published gene lists using defined criteria. For example, you can search for the SARS-CoV-2 interactome data from A549 cell by choosing Interactome in Technology and A549 in Source. See example below.
GO Heatmap shows the pre-computed Gene Ontology (GO) heatmap for the top GO terms for all the gene lists.
Browse All provides detailed information for each study and gene list, including author, title of the paper, link to the resource, technology and other useful information. You can select the gene lists, and add or append to reference lists.
If publicly available, the gene lists were downloaded from supplementary tables in the publications, then processed according to the criteria suggested by the publications. The gene lists were ordered by p value, and trimmed to 300 maximum genes. If there is no clear cutoff suggested in the publication, the genes were selected based on Log2FC>1 or <-1 and BH correct P<0.01. If there were too few genes selected based on this threshold, a relaxed threshold may be used and shown in the comment column of the gene list.